If you follow Dr. John Campbell on YouTube, you will note that on Saturday June 11, 2022 he talked about the COVID-19 Lab-Leak Hypothesis and how the new report on the origins of SARS-CoV-2 did not make sense (Image 1).
Image 1. More specifically, he talked about the WHO set up of the SAGO (Scientific Advisory Group for the Origins of Novel Pathogens) which rendered its first report on the origins of the COVID-19 pandemic SARS-CoV-2 virus on June 09, 2022.
https://cdn.who.int/media/docs/default-source/scientific-advisory-group-on-the-origins-of-novel-pathogens/sago-report-09062022.pdfb
Excerpt from Executive summary:
“Ancestral strains to SARS-CoV-2 have a zoonotic origin with the closest genetically related viruses being beta coronaviruses, identified in Rhinolophus bats in China in 2013 (96.1 %) and Laos in 2020 (96.8%).
However, neither the virus progenitors nor the natural/intermediate hosts or spill-over event to humans have been identified.”
Dr. Campbell is very skeptical about the conclusion that SARS-CoV-2 was a direct zoonotic infection from animals given that the chair of SAGO, Dr. Marietjie Venter disagrees with the notion that 96.8% identical RNA sequences of bat coronavirus with the Wuhan SARS-CoV-2 strain is sufficient to assume that the origins of the SARS-CoV-2 pandemic was a zoonotic transmission. As well, there is no other evidence for a closely matched coronavirus of animal origin for SARS-CoV-2, making zoonotic infection, an unlikely source of SARS-CoV-2.
Most notably China has refused to provide samples of Wuhan Institute of Virology (WIV) lab workers who were hospitalized in November 2019 with respiratory-like symptoms and have not allowed access to lab records or samples on what coronaviruses were being handled at WIV in the fall of 2019.
For more information on what scientists do and don’t know please see the 17 June 2021 article at Nature:
https://www.nature.com/articles/d41586-021-01529-3
Given the reluctance of the Chinese authorities to release samples or information about the bat coronaviruses studied at WIV in the fall of 2019 leaves little hope of resolving this important issue on the origins of SARS-CoV-2. On the other hand, new evidence is consistent moreso with the lab-leak hypothesis than a direct zoonotic event in terms of the source of the SARS-CoV-2 (Wuhan) pandemic virus, as explained below.
New Information Potentially Relevant to the Origins of SARS-CoV-2
In a recently submitted invited review entitled “ Trained immunity involving HERV-K102 activation promotes recovery from COVID-19 providing a new innate immunity vaccination paradigm against pandemic RNA viruses” consisting of 12,134 words with 151 references [1], it was suggested:
a) along with Vitamin D and the early interferon response, HERV-K102 mediated trained (innate) immunity may comprise the elusive ( “innate immunity”) correlate of protection against SARS-CoV-2 severity,
b) which incidentally, also accounts for optimal vitamin D protection against infection/severe disease and in part the amplification of the interferon and inflammatory response [2] (through an alternative pathway ie., the cGAS-STING interferon response via the spreading of HERV-K102 particles throughout the body and upon entry, the release of HERV-K102 cDNA genomes into the cytoplasm of cells),
c) the astonishing high level of SARS-CoV-2 mechanisms for targeting of HERV-K102 particle production and/or release in sebocytes (foamy macrophages constitutively producing HERV-K102 particles in the mucosa) in the upper respiratory tract (URT) [3] and lipid body negative foamy macrophages (LB-FMs) in the lower respiratory tract (LRT) [4]; including the interference of macrophage foam cell formation, the prevention of lytic release of the HERV-K102 particles from the LB-FMs by inducing apoptosis resistance, the conversion of the protector LB-FMs producing the protector HERV-K102 particles to long lived lipid body positive foamy macrophages (LB+FMs) producing high levels of SARS-CoV-2 virions (Image 2), etc.
not only validates HERV-K102 as the critical defense mechanism against severe COVID-19 but at the same time, strongly supports the lab-leak hypothesis.
Image 2. Antibody Dependent Enhancement (ADE) of Infection in the LB-FMs Converts the “Protector-Inflammatory” Foamy Macrophages to the “Progression-Mediating” LB+FMs the Latter Which Provides an Immunologically Privileged Site for SARS-CoV-2 Replication [5]
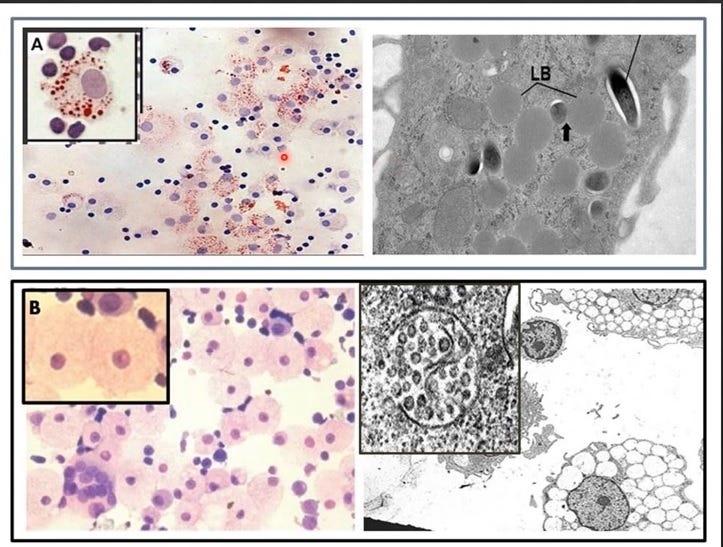
(see Image 4 below for H&E stains showing both types of foamy macrophages as induced by Mycobacterium tuberculosis)
First and foremost, selection for these traits could not have occurred in animals as only humans have the human endogenous retrovirus K102 (HERV-K102) protector foamy virus [6]. Indeed most species have their own version of foamy viruses. Therefore the only possibility of selection for traits inhibiting HERV-K102 particle-mediated defenses is through in vivo passage such as through humanized mice for example the BLT (Bone marrow, liver, thymus) humanized mouse. An improved version for studying respiratory viruses which includes human lung xenotransplants (the BLT-L mouse) was recently validated and coincidently published in October 2019 [7]. This paper includes authorship from an American coronavirus virologist, Dr. RS Baric, who has been linked to the WIV and gain of function research involving coronaviruses [https://www.technologyreview.com/2021/06/29/1027290/gain-of-function-risky-bat-virus-engineering-links-america-to-wuhan/].
From Ren et al [4], the myeloid cell types putatively producing HERV-K102 particles were identified as the Macro_c5-WDR74 subset and the progenitor as the Mono_c4-CD14-CD16 subset [4].
Note that release of protector particles from the foamy macrophages is by cell lysis which is why replenishment of foamy macrophages by the differentiation of committed monocytes (Mono_c4-CD14-CD16) is critical to recovery (and that unlike rodents, human macrophages do not replicate). According to Ren et al [4] both cell types are associated with recovery from moderate COVID-19 and are depleted with progression to more severe COVID-19. The same macrophage phenotype was shown in humanized mice mediating recovery from mild COVID-19 [2].
Interestingly, of all cell types in the lungs, both cell types uniquely express VDR [4], ie., the vitamin D receptor; and where it is well established that vitamin D deficiency associates with COVID-19 severity [8]. In foamy macrophages, vitamin D is needed to prevent conversion from lipid body negative (pro-inflammatory, M1-like) foamy macrophages (producing the foamy virus HERV-K102 particles) to the lipid body positive (anti-inflammatory, M2-like) foamy macrophages which do not produce the protector HERV-K102 particles (Image 2) [9]. This conversion is exacerbated in individuals with type 2 diabetes [9]. Note that vitamin D also blocks the induction of immune-responsive gene 1 (IRG1 genecard: aconitate decarboxylase 1, ACOD1) a very highly inflammatory protein induced by IRF-1 but blocked by functional VDR in macrophages [10].
These recovery-associated myeloid monocytes and foamy macrophages also both expressed:
a. IRF1 (needed for the induction of HERV-K102 proviral genome transcripts) [11]. Note that low IRF1 in PBMC monocytes at the time of SARS-CoV-2 diagnosis is a risk factor for COVID-19 severity and death [12];
b. CST3 (cytostatin C, an inhibitor of cysteine proteases and which interacts with C3, C4A, albumin, APP and alpha-fetoprotein (AFP) where the latter is required to prevent programmed cell death until it is time for the release of particles);
c. LYZ (an anti-microbial gene);
d. CEBPB and SPI1 (enhancers ascribed to trained (innate) immunity [13]). Also note that CEBPB is needed for the induction of PPARG for foam cell formation and is involved in the interferon gamma signaling pathway where IFNG induces HERV-K102 proviral transcripts [11]. Also SPI1 is involved in the activation and differentiation of macrophages or B cells, connected with HDAC1, a histone deacetylase with interacts with TP53 and RB1, SPI1 initiates transcription of the LTR of HIV-1 and directly interacts with IRF1;
e. IRF5 (needed for Type I IFN, IFNG and TLR signaling)
f. NR4A1 (needed for malignant phenotype known as EMT, FCER1 signaling and retroviral (HIV-1) lifecycle; pathway involves MAPK8, MAPK14, BCL2, TP53, RELA);
g. FOS and JUN (AP1 generic transcription factors);
h. CEBPD (involved in macrophage activation and differentiation and connects with HDAC1/3/4, SMAD3/4 and alpha-fetoprotein (AFP));
i. YBX1 (a cold shock protein involved in PI3K/AKT signaling needed for expression of HERV-K102 envelope protein, and which interacts with DDX6 needed for the transport of HERV-K102 proviral full-length transcripts for encapsidation into particles);
j. C1QBP and TP53 (both bind RNA and DNA, role in MHC Class II expression and DNA repair respectively);
k. ZEB2 (promotes EMT involved in TGFB1 signaling through SMADs);
l. POU2F2 (modulates pathways of FCER1, NR3C1- the glucocorticoid receptor, AR- the androgen receptor, and BCR-the B cell receptor and plays a role in retroviral (HIV-1) gene expression);
m. MAFB (represses ETS-1 erythroid specific genes and activates insulin and glucagon promoters);
n. KLR4/6/10 (kruppel-like receptors: KLR4 maintains embryonic stem cells and connects with PPARG and TP53, KLR6 is a tumor suppressor involved in the modulation of the aryl hydrocarbon receptor (AHR) and interacts with HIF1A (hypoxia response), RELA, NFKB1A, CXCL8, CCL2, and HDAC1/3, KLR10 regulates the circadian clock affecting genes involved in lipogenesis, glycolysis and TGFB1 signaling);
o. ARID5B (regulates adipogenesis);
p. ZNF688 and ZNF706.
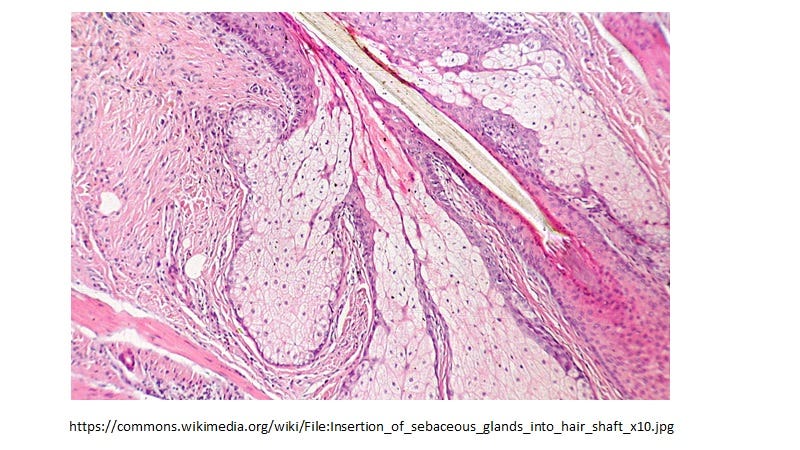
Sebocytes in sebaceous glands which line the mucosa, are specialized foamy macrophages constitutively producing and releasing HERV-K102 protector particles also by lysis [1]. This generates sebum which coats the mucosa positioning the HERV-K102 particles as a first line of defense. Sebocytes also express VDR and share all the differentially expressed transcription factors listed above which is common to foamy macrophages and the CD14+CD16+ monocyte progenitors. In other words the phenotype programming is the same in sebocytes as the LB-FMs and they have the identical morphology (Image 3 & Image 4, respectively).
Image 3. Sebocytes have the identical morphology to LB-FMs and upon programmed cell death secrete HERV-K102 particles in sebum which coats the mucosa.
Image 4. Two Types of Foamy Macrophages in Humans: A) the Lipid Body Positive Foamy Macrophages (LB+FMs, a Haven for Pathogens like Mycobacteria) [14] and B) the Lipid body Negative Foamy Macrophages (LB-FMs Producing the Protective HERV-K102 Foamy Virus Particles)[15,16]
Various Mechanisms by Which SARS-CoV-2 Infection of the Recovery Myeloid Cells Targets Trained Immunity/HERV-K102.
NOVEL ADE Mechanism Mediating Infection of the Recovery Myeloid Cells in the Lungs
In the lungs, progression to severe COVID-19 is associated with a novel type of antibody mediated enhancement (ADE) which mediates infection in macrophages and which has not seen before [4]. Neutralizing antibody to SARS-CoV-2 spike protein blocks the spike: ACE2 interaction (primary receptor), potentiating the spike: basigin (BSG or CD147) secondary receptor interaction (pre omicron strains). With the onset of COVID-19 severity in the lungs (correlated with the production of adaptive antibodies to spike protein especially the neutralizing antibodies), the infection switches from spike:ACE2 to spike:BSG. Cells expressing BSG in the lungs are primarily immune cells such as macrophages, monocytes, neutrophils, plasma B cells, T cells, NK cells, but also ciliated epithelial cells [4]. Thus, the infection of the recovery myeloid cells only occurs upon the generation of neutralizing antibodies to spike protein of SARS-CoV-2. Therefore, it is important for trained immunity/HERV-K102 to clear SARS-CoV-2 before adaptive immunity produces neutralizing, enhancing or other spike specific antibodies.
In autopsy materials from those who died from COVID-19, all the myeloid cell types in lungs were productively infected by SARS-CoV-2 [17].
Classical ADE in the URT for Sebocytes
In sebocytes, unlike the 4 other types of macrophages identified in nasopharyngeal swabs (NPS) [3], these “SARS-CoV-2 infected” cells did not express the trained immunity enhancers (SPI1, CEBPB), BSG (master regulator of foam cell formation and induction of inflammation), and showed the lowest levels of IFNGR1/2, all needed for the production of HERV-K102 particles. The phenotyping of these infected cells revealed a profile consistent with TLR2/4 stimulated sebocytes [18]. Infection of sebocytes was associated with an upregulation of FCGR2A known to mediate classical ADE and the expression of factors known to be involved in generating or associated with cytokine storm such as: NFKB1, TLR2/4, IDO1, and S100A9. Moreover, the high level of LAMP1 and MX1 but low level of DNAse2 [3] is consistent with the sebocytes being programmed for cell death but with apoptosis resistance [19]. Thus, it appears that SARS-CoV-2 infection of sebocytes blocks the lytic release of the HERV-K102 particles and inhibits HERV-K102 particle production. The former is evidence of SARS-CoV-2 mediating ‘immunosenescence’ [20] which is known to increase the risk of COVID-19 severity [reviewed in 1].
SARS-CoV-2 infection in the mouse does not recreate the pneumonia associated with COVID-19, but BSG humanized mice does [18]. This implicates human BSG in the generation of cytokine storm which was confirmed by the use of a specific antibody to human BSG which blocked cytokine storm in these humanized BSG mice [21]. Thus, as discussed above in the lungs, the switch of the entry molecules from the spike:ACE2 to spike:BSG inherently is associated with cytokine storm but MORE IMPORTANTLY, enables SARS-CoV-2 to directly inhibit the HERV-K102 protection system by infecting the recovery monocytes and macrophages which produce the HERV-K102 particles.
Image 5. Overview of Six General Mechanisms by Which SARS-CoV-2 May Target and Inhibit the HERV-K102 Innate Immunity Protector System
For more details and references, please see post at
In addition to the six general mechanisms cited above, of potential clinical significance, the glucocorticoid receptor (NR3C1) was only detected in virus negative neutrophils, macrophages, B cells and NK cells in the Ren et al study [4]. While NR3C1 is elevated in mild patients, it is downregulated with COVID-19 severity and is negatively associated with CXCL8 overexpression and the accumulation of dysfunctional neutrophils non-responsive to type I interferons [22]. HERV-K102 has glucocorticoid response elements in its LTRs [23], implying it may be induced by cortisol associated with mild COVID-19 and may be downregulated associated with lower NR3C1 found in severe COVID-19. Thus, this illustrates yet another mechanism by which SARS-CoV-2 may inhibit the HERV-K102 protector system upon entry into the recovery myeloid cells.
As well Ren et al [4] reported that SARS-CoV-2 infected myeloid cells upregulated PPARA which would tend to convert the lipid body negative, proinflammatory, M1-like foamy macrophages (producing HERV-K102 particles) to the lipid body positive, anti-inflammatory, M2-like foamy macrophages; shutting down the production of HERV-K102 particles (Image 2).
All in all, this new suggestion that the HERV-K102 protector system correlates with protection against COVID-19 severity and that SARS-CoV-2 has been selected for traits which block or inhibit the HERV-K102 protector system, argues that selection could have only occurred within a milieu involving human immune cells in vivo such as serial passage of a bat coronavirus through the BLT or BLT-L humanized mouse [7].
It is unclear if Dr. Fauci’s definition of ‘gain of function’ research included serial passage of coronaviruses through humanized BLT or BLT-L mice rather than reverse genetics or recombinant forms of coronaviruses.
Vaccines Provided Selection Pressure for the Generation of SARS-CoV-2 Variants
Prior to the introduction of the COVID-19 vaccines, there was not much in terms of selection of immune escape variants [1] which argues that selection of an optimally fit SARS-CoV-2 for humans characterized the original Wuhan strain. Indeed, the WHO suggested the SARS-CoV-2 pandemic virus genome was stable earlier in the pandemic, but following the widespread introduction of the COVID-19 vaccines, they then declared a problem with the selection of variants.
Surprisingly, Little Natural Selection Pressure for Generation of Variants
Rather contrary to expectations, selection of variants were not commonly characterized in immunocompromised patients with comorbidities hospitalized for extended periods of time but instead occurred (on average about 10 days) when monoclonal neutralizing antibodies such as bamlanivimab to the spike protein were clinically used in these patients [24]. Similar problems were reported with use of convalescent plasma [25]. Indeed, given the unexpected mechanism of switch from spike:ACE2 primary receptor to spike:BSG secondary receptor associated with the onset of neutralizing antibodies, means there was very little selection pressure for generating SARS-CoV-2 variants in the lungs associated with severity in vivo in humans during natural infection.
Vaccination Produced Selection Pressure in the Oral-Nasopharyngeal Mucosa (ie. for Infection)
Instead as reviewed in reference 1 along with new convincing data for Canada for the selection of the alpha then delta variants, COVID-19 full vaccination (where the second dose generated significant levels of neutralizing/enhancing antibodies) was largely responsible for the selection of variants. This point was proven in reference [26] that the COVID-19 vaccines selected for variants. This selection commonly occurred in the oral-nasopharyngeal mucosa and likely involved classical ADE mechanisms associated with the upregulation of FCGR2A in sebocytes [3] resulting in higher infection rates for the selected variants in vaccinated over non-vaccinated individuals [1].
One Dose Immunization Superior to Two Dose Vaccination: Similar VEs but No Selection Pressure for Variants and Evidence for a 60 to 70 % Reduction in Non-COVID-19 All Cause Mortality
It is noteworthy that in clinical trials or test negative design studies, most of the transmissions following the first dose appeared to have occurred at vaccination clinics [27 and reviewed in 1]. Thus, ironically, vaccination introduced ‘iatrogenic’ disease particularly after dose 1. This artificially reduced the ‘vaccine effectiveness (VE) of the first dose. This is important as the VE following the first dose after 33 days was instead calculated to be about 91 % [27] on par with a full two dose immunization [1]. The single dose of mRNA vaccines induced trained immunity putatively involving HERV-K102 as evidenced by the heterologous protection against non-COVID-19 mortality of about 60 to 70 % which was not impacted by administration of the second dose [28].
However, subsequent SARS-CoV-2 infection eliminated the beneficial heterologous protection against non-COVID-19 mortality. This was shown for Canadians based on the loss of negative all cause excess mortality due to neutralizing antibodies produced by the second dose and presumed ADE. The latter was associated with the initial selection and/or full emergence (ie., greater than 50% of the variants) of the alpha and delta variants in Canada [1, 29].
Thus, on closer inspection, one dose of the mRNA COVID-19 vaccines in all likelihood would have provided the protection needed for INNATE MEDIATED herd immunity (Image 6, February 2021 ONS data) without the selection of variants and which conceivably would have ended the pandemic sooner.
Image 6. The Ratio of Mortality Rates in the Ever Vaccinated over the Never Vaccinated Reveals only One Dose (February 2021 at about 96 % of the ever vaccinated, mostly >50+) Provided Protection
Summary and Conclusions
Thus, it seems quite plausible that the original Wuhan strain had already undergone selection by the human immune system prior to its inadvertent ‘accidental’ release in fall of 2019. This conclusion was also reached by as early as May 2020 based on the sequencing of SARS-CoV-2 compared with SARS-CoV-1 over the first few months of spread in humans [30].
This new review [1] provides the rationale to suggest in contrast to the first report of SAGO released June 9, 2022, that the lab-leak hypothesis is instead, the most likely source of the SARS-CoV-2 pandemic virus.
REFERENCES
Laderoute MP. Trained immunity involving HERV-K102 activation promotes recovery from COVID-19 providing a new innate immunity vaccination paradigm against pandemic RNA viruses. (submitted review).
Kenney DJ, O'Connell AK, Turcinovic J, et al. Humanized mice reveal a macrophage-enriched gene signature defining human lung tissue protection during SARS-CoV-2 infection. Cell Rep. 2022 Apr 19;39(3):110714. doi: 10.1016/j.celrep.2022.110714.
Ziegler CGK, et al. Impaired local intrinsic immunity to SARS-CoV-2 infection in severe COVID-19. Cell. 2021 Sep 2;184(18):4713-4733.e22. doi: 10.1016/j.cell.2021.07.023.
Ren X, et al. COVID-19 immune features revealed by a large-scale single-cell transcriptome atlas. Cell. 2021 Nov 11;184(23):5838. doi: 10.1016/j.cell.2021.10.023.
Dias SSG, Soares VC, Ferreira AC, et al. Lipid droplets fuel SARS-CoV-2 replication and production of inflammatory mediators. PLoS Pathog. 2020 Dec 16;16(12):e1009127. doi: 10.1371/journal.ppat.1009127.
Subramanian RP, Wildschutte JH, Russo C, Coffin JM. Identification, characterization, and comparative genomic distribution of the HERV-K (HML-2) group of human endogenous retroviruses. Retrovirology. 2011 Nov 8;8:90. doi: 10.1186/1742-4690-8-90.
Wahl A, De C, Abad Fernandez M, Lenarcic EM, Xu Y, Cockrell AS, Cleary RA, Johnson CE, Schramm NJ, Rank LM, Newsome IG, Vincent HA, Sanders W, Aguilera-Sandoval CR, Boone A, Hildebrand WH, Dayton PA, Baric RS, Pickles RJ, Braunstein M, Moorman NJ, Goonetilleke N, Victor Garcia J. Precision mouse models with expanded tropism for human pathogens. Nat Biotechnol. 2019 Oct;37(10):1163-1173. doi: 10.1038/s41587-019-0225-9.
Jordan T, Siuka D, Rotovnik NK, Pfeifer M. COVID-19 and Vitamin D- a Systematic Review. Zdr Varst. 2022 Mar 21;61(2):124-132. doi: 10.2478/sjph-2022-0017.
Oh J, Weng S, Felton SK, Bhandare S, Riek A, Butler B, Proctor BM, Petty M, Chen Z, Schechtman KB, Bernal-Mizrachi L, Bernal-Mizrachi C. 1,25(OH)2 vitamin d inhibits foam cell formation and suppresses macrophage cholesterol uptake in patients with type 2 diabetes mellitus. Circulation. 2009 Aug 25;120(8):687-98. doi: 10.1161/CIRCULATIONAHA.109.856070.
Tallam A, Perumal TM, Antony PM, Jäger C, Fritz JV, Vallar L, Balling R, Del Sol A, Michelucci A. Gene Regulatory Network Inference of Immunoresponsive Gene 1 (IRG1) Identifies Interferon Regulatory Factor 1 (IRF1) as Its Transcriptional Regulator in Mammalian Macrophages. PLoS One. 2016 Feb 12;11(2):e0149050. doi: 10.1371/journal.pone.0149050.
Manghera M, Ferguson-Parry J, Lin R, Douville RN. NF-κB and IRF1 Induce Endogenous Retrovirus K Expression via Interferon-Stimulated Response Elements in Its 5' Long Terminal Repeat. J Virol. 2016 Sep 29;90(20):9338-49. doi: 10.1128/JVI.01503-16.
Utrero-Rico A, et al. Alterations in Circulating Monocytes Predict COVID-19 Severity and Include Chromatin Modifications Still Detectable Six Months after Recovery. Biomedicines. 2021 Sep 17;9(9):1253. doi: 10.3390/biomedicines9091253.
Arts RJW, Joosten LAB, Netea MG. The Potential Role of Trained Immunity in Autoimmune and Autoinflammatory Disorders. Front Immunol. 2018 Feb 20;9:298. doi: 10.3389/fimmu.2018.00298.
Peyron P, Vaubourgeix J, Poquet Y, et al. Foamy macrophages from tuberculous patients' granulomas constitute a nutrient-rich reservoir for M. tuberculosis persistence. PLoS Pathog. 2008 Nov;4(11):e1000204. doi: 10.1371/journal.ppat.1000204.
Laderoute MP, Giulivi A, Larocque L, et al. The replicative activity of human endogenous retrovirus K102 (HERV-K102) with HIV viremia. AIDS. 2007 Nov 30;21(18):2417-24.
Laderoute MP, Larocque LJ, Giulivi A, Diaz-Mitoma F. Further evidence that human endogenous retrovirus K102 is a replication competent foamy virus that may antagonize HIV-1 replication. Open AIDS J. 2015 Dec 7;9:112-22. doi: 10.2174/1874613601509010112.
Delorey TM, et al. COVID-19 tissue atlases reveal SARS-CoV-2 pathology and cellular targets. Nature. 2021 Jul;595(7865):107-113. doi: 10.1038/s41586-021-03570-8.
Torocsik D et al. Genome wide analysis of TLR1/2- and TLR4-activated SZ95 sebocytes reveals a complex immune-competence and identifies serum amyloid A as a marker for activated sebaceous glands. PLoS One. 2018 Jun 21;13(6):e0198323. doi: 10.1371/journal.pone.0198323.
Fischer H, Fumicz J, Rossiter H, Napirei M, Buchberger M, Tschachler E, Eckhart L. Holocrine Secretion of Sebum Is a Unique DNase2-Dependent Mode of Programmed Cell Death. J Invest Dermatol. 2017 Mar;137(3):587-594. doi: 10.1016/j.jid.2016.10.017.
Laderoute MP. A new paradigm about HERV-K102 particle production and blocked release to explain cortisol mediated immunosenescence and age-associated risk of chronic disease. Discov Med. 2015 Dec;20(112):379-91.
Geng J, et al. CD147 antibody specifically and effectively inhibits infection and cytokine storm of SARS-CoV-2 and its variants delta, alpha, beta, and gamma. Signal Transduct Target Ther. 2021 Sep 25;6(1):347. doi: 10.1038/s41392-021-00760-8.
Park JH, Lee HK. Re-analysis of Single Cell Transcriptome Reveals That the NR3C1-CXCL8-Neutrophil Axis Determines the Severity of COVID-19. Front Immunol. 2020 Aug 28;11:2145. doi: 10.3389/fimmu.2020.02145.
Ono M, Yasunaga T, Miyata T, Ushikubo H. Nucleotide sequence of human endogenous retrovirus genome related to the mouse mammary tumor virus genome. J Virol. 1986 Nov;60(2):589-98. doi: 10.1128/JVI.60.2.589-598.1986.
Jensen B, Luebke N, Feldt T, Keitel V, Brandenburger T, Kindgen-Milles D, Lutterbeck M, Freise NF, Schoeler D, Haas R, Dilthey A, Adams O, Walker A, Timm J, Luedde T. Emergence of the E484K mutation in SARS-COV-2-infected immunocompromised patients treated with bamlanivimab in Germany. Lancet Reg Health Eur. 2021 Sep;8:100164. doi: 10.1016/j.lanepe.2021.100164.
Kemp SA, Collier DA, Datir RP, Ferreira IATM, Gayed S, Jahun A, et al. SARS-CoV-2 evolution during treatment of chronic infection. Nature. 2021 Apr;592(7853):277-282. doi: 10.1038/s41586-021-03291-y.
Micochova P, Kemp SA, Dhar MS, Papa G, Meng B, Ferreira IATM, et al. SARS-CoV-2 B.1.617.2 Delta variant replication and immune evasion. Nature. 2021 Nov;599(7883):114-119. doi: 10.1038/s41586-021-03944-y.
Chung H, et al; Canadian Immunization Research Network (CIRN) Provincial Collaborative Network (PCN) Investigators. Effectiveness of BNT162b2 and mRNA-1273 covid-19 vaccines against symptomatic SARS-CoV-2 infection and severe covid-19 outcomes in Ontario, Canada: test negative design study. BMJ. 2021 Aug 20;374:n1943. doi: 10.1136/bmj.n1943.
Xu S, et al. COVID-19 Vaccination and Non-COVID-19 Mortality Risk - Seven Integrated Health Care Organizations, United States, December 14, 2020-July 31, 2021. MMWR Morb Mortal Wkly Rep. 2021 Oct 29;70(43):1520-1524. doi: 10.15585/mmwr.mm7043e2.
Laderoute MP. What might excess all cause mortality rates tell us about the emergence of SARS-CoV-2 variants, and overall effects of vaccines on mortality during the pandemic in Canada? February 12, 2022. hervk102.substack.com.
Zhan SH et al. SARS-CoV-2 is well adapted for humans. What does this mean for re-emergence? BioRxiv May 2 2020 (preprint). https://www.biorxiv.org/content/10.1101/2020.05.01.073262v1









Hi Dr. Marian,
I've checked reference 4
"The presence of SARS-CoV-2
RNA in various immune cell types, including neutrophils, macro-
phages, plasma B cells, T cells, and NK cells, was surprising to
us initially, but the research community is beginning to appre-
ciate this phenomenon. While it is still not clear how such im-
mune cells would acquire viral sequences..."
and given this two facts from the literature:
*SARS-CoV-2 invades host cells via a novel route: CD147-spike protein
https://www.biorxiv.org/content/10.1101/2020.03.14.988345v1
*"CD147, a transmembrane glycoprotein, is expressed on all leukocytes, platelets, and endothelial cells." https://pubmed.ncbi.nlm.nih.gov/24372217/
Your conclusion here is very interesting:
"Thus, the infection of the recovery myeloid cells only occurs upon the generation of neutralizing antibodies to spike protein of SARS-CoV-2. Therefore, it is important for trained immunity/HERV-K102 to clear SARS-CoV-2 before adaptive immunity produces neutralizing antibodies."
Thank you for sharing!
Take care,
Agus
Others have noted that SARS-CoV-2 appears to have been preadapted for human transmission. Zhan SH et al, BioRxiv May 2 2020 (preprint). https://www.biorxiv.org/content/10.1101/2020.05.01.073262v1
“By the time SARS-CoV-2 was first detected in late 2019, it was already pre-adapted to human transmission to an extent similar to late epidemic SARS-CoV.”
Other sources of information include:
https://jamiemetzl.com/origins-of-sars-cov-2/
https://nicholaswade.medium.com/origin-of-covid-following-the-clues-6f03564c038
https://www.vanityfair.com/news/2021/06/the-lab-leak-theory-inside-the-fight-to-uncover-covid-19s-origins